My name is Letizia Lo Faro and I am a Post-Doctoral research scientist in the Oxford Transplant research group. I have so far used QUOD samples in 4 or 5 separate research studies. In one of these, conducted together with Ms Flavia Neri (University of Padova), we were interested in studying the molecular features of donor kidneys with acute kidney injury (AKI) having different functional outcomes after transplant.

Once we fully characterised our research question, we moved onto donor sample selection. We decided to compare 4 different groups of donors: with or without AKI and each with good (≥45 mL/min) or poor (<45 mL/min) function (eGFR) 12 months post-transplant, for a total of 40 donors. We maximised the difference between outcomes as much as possible and to allow for fair comparisons we decided to match the groups for several other variables (donor age and gender, BMI, cold ischaemia time, recipient age, recipient gender…just to name a few). I believe appropriate sample selection is a key step in such retrospective studies and we engaged with the QUOD Data Manager earlier on in the process. This provided us with a great overview of all the variables available and also made sure our groups were nicely balanced.
Once we were happy with the donor selection, we proceeded with requesting the samples from the biobank. In this case they were full RNAlater frozen kidney biopsies and formalin-fixed paraffin-embedded (FFPE) kidney tissue slides. In this study we were interested in studying protein expression, so first of all we processed the tissue biopsies to mechanically homogenise them and extract the proteins. Later on the proteins were quantified and the expression of 17 proteins of interest was analysed by Western Blotting (a technique where a mixture of proteins is separated on a gel, based on the molecular weight, and, following transfer on a membrane, proteins of interest are identified by binding to specific antibodies). Results were then analysed with statistical tools.
A set of FFPE slides was then utilised for histological assessment, by standard staining with haematoxylin and eosin (H&E), to quantify chronic and acute tissue damage. One additional set of slides was utilised for confirmation of the western blotting results by immunohistochemistry, another method which allows to check for the presence of certain proteins/products in a tissue, by binding with labelled antibodies.
Results from this study were presented at the British Transplantation Society meeting last year and we are currently finalising a manuscript for publication.
Our results suggested that specific molecular patterns are recognizable in acutely inured kidneys that proceed towards worse function after transplantation. In particular, Peroxisome proliferator-activated receptor gamma (PPAR gamma) specifically increased after acute injury that progressed to worse function, underlining a potential role of metabolic dysfunction in the development of kidney disease (Figure 1).
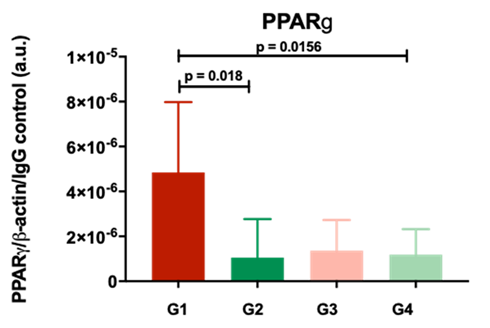

Figure 1. The protein PPARg, quantified by western blotting analysis, is significantly increased in AKI kidneys with poor outcomes 12months post-transplant.
Our findings have helped identify potential molecular mechanisms involved in the progression of acute kidney injury to chronic kidney disease and post-transplant dysfunction and may constitute a therapeutic target of further interventions aimed at improving the quality of donated kidneys with an acute injury.
Working with QUOD samples has allowed us to really dive deep into the study of molecular mechanisms of kidney injury and the availability of different sample types also allowed us to apply various techniques and methods to validate our findings.
